3.2 Hierarchical Clustering
3.2 Hierarchical Clustering
3.2.1 Dendrogram for Small Data Set
3.2.1 Dendrogram for Small Data Set
In[]:=
session=StartExternalSession["Python"]
Out[]=
ExternalSessionObject
Test
In[]:=
5+6
Out[]=
11
In[]:=
from sklearn.datasets import make_blobs
from scipy.cluster.hierarchy import linkage
import numpy as np
from sklearn.cluster import AgglomerativeClustering
from scipy.cluster.hierarchy import linkage
import numpy as np
from sklearn.cluster import AgglomerativeClustering
In[]:=
X, y = make_blobs(random_state=1)
In[]:=
X
Out[]=
NumericArray
In[]:=
Normal[%]
Out[]=
{{-0.794152,2.10495},{-9.15155,-4.81286},{-11.4418,-4.45781},{-9.76762,-3.19134},{-4.53656,-8.40186},{-6.26302,-8.10666},{-6.38481,-8.47303},{-9.20491,-4.57688},{-2.76018,5.55121},{-1.17104,4.33092},{-10.0364,-5.56912},{-9.87589,-2.82386},{-7.17533,-8.77059},{-2.40672,6.09894},{-4.87418,-10.0496},{-6.07855,-7.93969},{-6.83239,-7.47068},{-2.34673,3.56128},{-10.3416,-3.90975},{-11.0926,-3.78397},{-6.50212,-7.91249},{-10.2639,-3.92073},{-6.81608,-8.44987},{-1.34052,4.15712},{-10.373,-4.59208},{-7.375,-10.5881},{-6.62352,-8.25338},{-1.35939,4.05424},{-0.197452,2.34635},{-6.54431,-9.29757},{-1.92745,4.93685},{-2.80208,4.05715},{-7.58198,-9.15025},{-1.8514,3.51886},{-8.37006,-3.61534},{-7.25145,-8.25497},{-8.79879,-3.76819},{-11.3708,-3.63819},{-10.1786,-4.55727},{-7.20133,-8.27228},{-6.78422,-8.22634},{-9.64717,-5.26563},{-1.98198,4.02244},{-11.2278,-3.40281},{-9.79941,-3.83434},{-6.53542,-8.01553},{-0.757969,4.90898},{0.526016,3.00999},{-2.77687,4.64091},{-1.78245,3.47072},{-10.22,-4.15411},{-6.40583,-9.78067},{-6.98706,-7.53485},{-7.46576,-7.32922},{-1.5394,5.02369},{-6.56967,-8.32793},{-10.6177,-3.25532},{-8.72396,-1.98625},{-1.61735,4.98931},{-1.14663,4.1084},{-9.81115,-3.5433},{-7.7118,-7.25174},{-6.5617,-6.86},{-10.0223,-4.72851},{-11.8557,-2.71718},{-5.73343,-8.44054},{-2.41396,5.65936},{-8.33744,-7.83968},{-1.83199,3.52863},{-9.57422,-3.87601},{-9.59422,-3.35977},{-9.25716,-4.90705},{-6.46256,-7.73295},{-0.820576,5.33759},{0.000242271,5.14853},{-9.68208,-5.97555},{-6.196,-7.40282},{-7.02121,-8.37954},{-2.18773,3.33352},{-10.4448,-2.72884},{-0.527931,5.92631},{-11.197,-3.09},{-9.83768,-3.07718},{-5.16022,-7.04217},{-2.35122,4.00974},{-0.52579,3.3066},{-1.46864,6.50675},{-0.758704,3.72276},{-10.3039,-3.12537},{-2.33081,4.39383},{-5.90454,-7.78374},{-1.60875,3.76949},{-1.86845,4.99311},{-10.6684,-3.57578},{-8.87629,-3.54445},{-6.02606,-5.96625},{-7.04747,-9.27525},{-1.37397,5.29163},{-6.25393,-7.10879},{0.0852519,3.64528}}
In[]:=
datap=%;
In[]:=
Needs["HierarchicalClustering`"]
In[]:=
DendrogramPlot[datap,TruncateDendrogram18,HighlightLevel3,OrientationLeft]
Out[]=
In[]:=
X, y = make_blobs(random_state=1)
ac = AgglomerativeClustering(n_clusters=3,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
ac = AgglomerativeClustering(n_clusters=3,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
Out[]=
NumericArray
In[]:=
Normal[%]
Out[]=
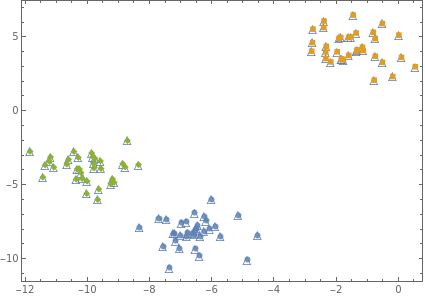
{1,2,2,2,0,0,0,2,1,1,2,2,0,1,0,0,0,1,2,2,0,2,0,1,2,0,0,1,1,0,1,1,0,1,2,0,2,2,2,0,0,2,1,2,2,0,1,1,1,1,2,0,0,0,1,0,2,2,1,1,2,0,0,2,2,0,1,0,1,2,2,2,0,1,1,2,0,0,1,2,1,2,2,0,1,1,1,1,2,1,0,1,1,2,2,0,0,1,0,1}
In[]:=
labels=%;
In[]:=
p0=ListPlot[datap,Frame->True,PlotMarkers"",AxesNone]
Out[]=
In[]:=
dataC=MapThread[Flatten[{#1,#2}]&,{datap,labels}];
In[]:=
dataC1=Map[{#[[1]],#[[2]]}&,Select[dataC,#[[3]]0&]];
In[]:=
dataC2=Map[{#[[1]],#[[2]]}&,Select[dataC,#[[3]]1&]];
In[]:=
dataC3=Map[{#[[1]],#[[2]]}&,Select[dataC,#[[3]]2&]];
In[]:=
Show[{p0,ListPlot[{dataC1,dataC2,dataC3},Frame->True,PlotMarkersAutomatic,AxesNone]},AspectRatio0.7]
Out[]=
In[]:=
from sklearn import metrics
s=metrics.silhouette_score(X,prediction,'euclidean')
s
s=metrics.silhouette_score(X,prediction,'euclidean')
s
Out[]=
0.769709
In[]:=
DendrogramPlot[datap,TruncateDendrogram18,HighlightLevel2,OrientationLeft]
Out[]=
In[]:=
X, y = make_blobs(random_state=1)
ac = AgglomerativeClustering(n_clusters=2,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
ac = AgglomerativeClustering(n_clusters=2,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
Out[]=
NumericArray
In[]:=
Normal[%]
Out[]=
{1,0,0,0,0,0,0,0,1,1,0,0,0,1,0,0,0,1,0,0,0,0,0,1,0,0,0,1,1,0,1,1,0,1,0,0,0,0,0,0,0,0,1,0,0,0,1,1,1,1,0,0,0,0,1,0,0,0,1,1,0,0,0,0,0,0,1,0,1,0,0,0,0,1,1,0,0,0,1,0,1,0,0,0,1,1,1,1,0,1,0,1,1,0,0,0,0,1,0,1}
In[]:=
labels=%;
In[]:=
dataC=MapThread[Flatten[{#1,#2}]&,{datap,labels}];
In[]:=
dataC1=Map[{#[[1]],#[[2]]}&,Select[dataC,#[[3]]0&]];
In[]:=
from sklearn import metrics
s=metrics.silhouette_score(X,prediction,'euclidean')
s
s=metrics.silhouette_score(X,prediction,'euclidean')
s
In[]:=
X, y = make_blobs(random_state=1)
ac = AgglomerativeClustering(n_clusters=4,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
ac = AgglomerativeClustering(n_clusters=4,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
In[]:=
from sklearn import metrics
s=metrics.silhouette_score(X,prediction,'euclidean')
s
s=metrics.silhouette_score(X,prediction,'euclidean')
s
3.2.2 Image Segmentation
3.2.2 Image Segmentation
In[]:=
from numpy import array, matrix
from scipy.io import mmread, mmwrite
import numpy as np
from numpy import array, matrix
from scipy.io import mmread, mmwrite
import numpy as np
In[]:=
from scipy.cluster.hierarchy import linkage
import numpy as np
from sklearn.cluster import AgglomerativeClustering
import numpy as np
from sklearn.cluster import AgglomerativeClustering
In[]:=
X=mmread('dataX.mtx')
In[]:=
ac = AgglomerativeClustering(n_clusters=3,affinity='euclidean',linkage='complete')
labels = ac.fit_predict(X)
prediction=labels
prediction
labels = ac.fit_predict(X)
prediction=labels
prediction