4.5 Symbolic Regression Models
4.5 Symbolic Regression Models
4.5.1 Model with Single Variable
4.5.1 Model with Single Variable
In[]:=
session=StartExternalSession["Python"]
Out[]=
ExternalSessionObject
Test
In[]:=
5+6
Out[]=
11
In[]:=
SeedRandom[3512]
In[]:=
data=Table[{x,Sin[2x]+xExp[Cos[x]]+RandomVariate[NormalDistribution[0,1.5]]},{x,RandomReal[{-10,10},500]}];
In[]:=
p0=ListPlot[data]
Out[]=
In[]:=
fit=FindFormula[data,x,10000,All,PerformanceGoal->"Quality",SpecificityGoal1];
In[]:=
fitN=fit//Normal//Normal;
In[]:=
fitN[[1]]
Out[]=
1.27351x+1.08506xCos[x]Score-1.7471,Error4.15346,Complexity13
In[]:=
fitN[[1]][[2]][[2]]
Out[]=
4.15346
In[]:=
fitN[[1]][[2]][[3]]
Out[]=
13
In[]:=
Candidates=Map[{#[[2]][[2]],#[[2]][[3]]}&,fitN]
Out[]=
{{4.15346,13},{4.27686,13},{4.30271,13},{4.14871,14},{4.17143,14},{4.28354,13},{4.30785,13},{4.24282,14},{4.46804,13},{4.35183,14},{4.78672,13},{3.40504,27},{3.40509,27},{3.39193,28},{3.39197,28},{3.46518,28},{5.17748,13},{3.87729,27},{3.84902,28},{3.85578,28},{4.05993,27},{4.06316,27},{2.54501,46},{4.22627,31},{4.244,31},{4.24915,31},{4.25661,31},{4.44057,30},{4.33227,31},{4.44438,30},{4.44597,30},{4.46072,30},{4.64682,30},{5.31634,30},{5.33443,30},{6.41688,30},{24.7371,1},{24.6164,4},{24.6212,4},{24.7322,4},{24.7327,4},{24.7391,4},{25.0854,4},{24.6104,7},{24.623,7},{24.6317,7},{24.6333,7},{24.646,7},{24.8078,7},{61.4789,1}}
In[]:=
Length[Candidates]
Out[]=
50
In[]:=
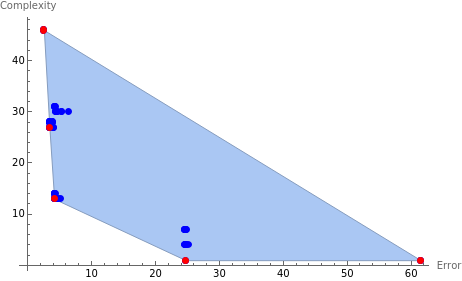
p2=ListPlot[Candidates,PlotStyle{PointSize[0.015],Blue},AxesLabel{"Error","Complexity"}];
In[]:=
cH=ConvexHullMesh[Candidates];
In[]:=
ParetoFront=MeshCoordinates[RegionBoundary[cH]]
Out[]=
{{4.15346,13.},{3.40504,27.},{2.54501,46.},{24.7371,1.},{61.4789,1.}}
In[]:=
p1=ListPlot[ParetoFront,PlotStyle{PointSize[0.015],Red}];
In[]:=
Show[{p2,p1,cH,p2,p1}]
Out[]=
In[]:=
Map[Norm[{{0,0}-#},1]&,ParetoFront]
Out[]=
{13.,27.,46.,24.7371,61.4789}
In[]:=
Norm[{{0,0}-ParetoFront[[2]]},1]
Out[]=
27.
In[]:=
modelOptimal=Select[fitN,{#[[2]][[2]],#[[2]][[3]]}ParetoFront[[2]]&]
Out[]=
{0.945963xCos[x]+9.34452Sin[0.182589x]+1.93301Sin[2.14391x]Score-1.89644,Error3.40504,Complexity27}
In[]:=
fitOpt=(modelOptimal//Normal)[[1,1]]
Out[]=
0.945963xCos[x]+9.34452Sin[0.182589x]+1.93301Sin[2.14391x]
In[]:=
Show[{p0,Plot[fitOpt,{x,-10,10},PlotStyle{Thin,Red}]}]
Out[]=
In[]:=
modelHighAccuracy=Select[fitN,{#[[2]][[2]],#[[2]][[3]]}ParetoFront[[3]]&]
Out[]=
{-0.0291352+0.195613xCos[x]+9.5328Sin[0.124476x]+3.47887Cos[x]Sin[1.1323x]+6.88565Cos[x]Sin[0.158737(0.173567+x)]Score-2.07762,Error2.54501,Complexity46}
In[]:=
fitHigh=(modelHighAccuracy//Normal)[[1,1]]
Out[]=
-0.0291352+0.195613xCos[x]+9.5328Sin[0.124476x]+3.47887Cos[x]Sin[1.1323x]+6.88565Cos[x]Sin[0.158737(0.173567+x)]
In[]:=
p1=Plot[fitHigh,{x,-10,10},PlotStyle{Thin,Red}];
In[]:=
Show[{p0,p1}]
Out[]=
4.5.2 Surface Fitting
4.5.2 Surface Fitting
In[]:=
from numpy import array, matrix
from scipy.io import mmread, mmwrite
from math import exp, sin, cos
function_set = ['add', 'sub', 'mul', 'div',
'sqrt', 'log', 'abs', 'neg', 'inv',
'max', 'min','sin','cos','exp']
from numpy import array, matrix
from scipy.io import mmread, mmwrite
from math import exp, sin, cos
function_set = ['add', 'sub', 'mul', 'div',
'sqrt', 'log', 'abs', 'neg', 'inv',
'max', 'min','sin','cos','exp']
In[]:=
from gplearn.genetic import SymbolicRegressor
import numpy as np
from sklearn.utils.random import check_random_state
import numpy as np
from sklearn.utils.random import check_random_state
In[]:=
x0 = np.arange(-1, 1, 1/10.)
x1 = np.arange(-1, 1, 1/10.)
x0, x1 = np.meshgrid(x0, x1)
y_truth = x0**2 - x1**2 + x1 - 1
x1 = np.arange(-1, 1, 1/10.)
x0, x1 = np.meshgrid(x0, x1)
y_truth = x0**2 - x1**2 + x1 - 1
In[]:=
rng = check_random_state(0)
X_train = rng.uniform(-1, 1, 100).reshape(50, 2)
y_train = X_train[:, 0]**2 - X_train[:, 1]**2 + X_train[:, 1] - 1
X_train = rng.uniform(-1, 1, 100).reshape(50, 2)
y_train = X_train[:, 0]**2 - X_train[:, 1]**2 + X_train[:, 1] - 1
In[]:=
est_gp = SymbolicRegressor(population_size=5000,
generations=20, stopping_criteria=0.01,
p_crossover=0.7, p_subtree_mutation=0.1,
p_hoist_mutation=0.05, p_point_mutation=0.1,
max_samples=0.9, verbose=1,
parsimony_coefficient=0.01, random_state=0).fit(X_train, y_train)
generations=20, stopping_criteria=0.01,
p_crossover=0.7, p_subtree_mutation=0.1,
p_hoist_mutation=0.05, p_point_mutation=0.1,
max_samples=0.9, verbose=1,
parsimony_coefficient=0.01, random_state=0).fit(X_train, y_train)
In[]:=
print(est_gp._program)