GTAtomsInCell
GTAtomsInCell[object,cluster,plotdata]
finds all atoms in a region defined by object in cluster. plotdata is used to customize the plot.
Details and Options
- If a larger cluster, or a complicated structure is constructed it is sometimes hard to inspect the local arrangement of the atoms. In the standard construction of the cluster all atoms inside a sphere are found. With this command we can cut out of cluster the atoms in a certain region defined by object. In the moment the region is a parallelepiped. plotdata helps to customize the plot. It is a list {distance,size}, where distance is the maximum bond length used to plot bonds and size is a parameter for the size of the atoms.
- The following options can be given:
-
GOPlot True Decides if the result is plotted GOVerbose True Decides if additional information is provided. - This command is not explained in: W. Hergert, M. Geilhufe, Group Theory in Solid State Physics and Photonics. Problem Solving with Mathematica
Examples
open allclose allBasic Examples (2)
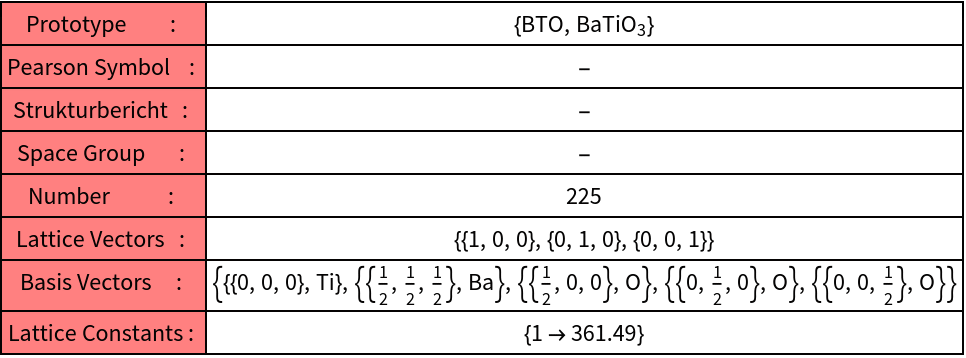
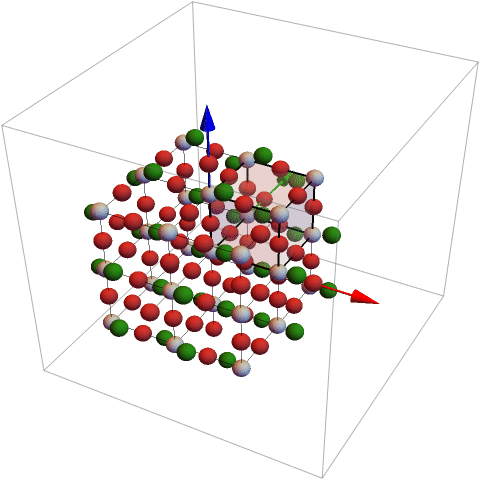
From the dataset the structure of barium titanate is extracted
A graphics object is defined from the parallelepiped and showed finally together with the cluster.
The atoms will be found in the region:
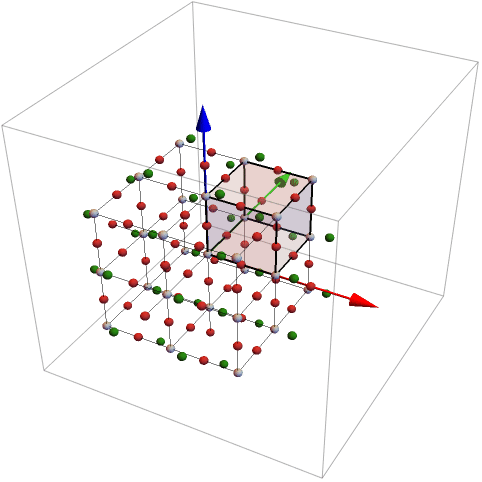
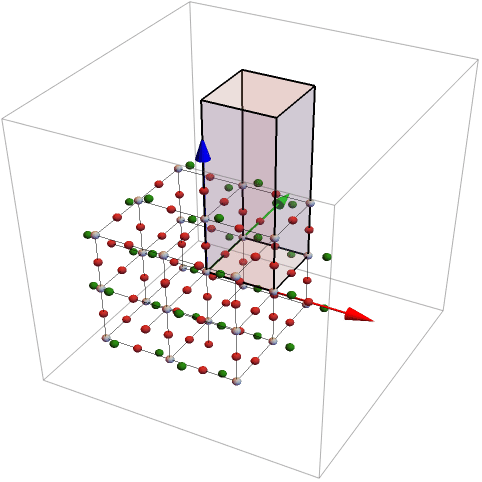
The region can be defined in different ways. The following list defines also a parallelepiped. The last argument in the last can be used to rotate the object
For the definition of the rotation matrix two vectors are given. The first vector is rotated into the second.
Options (2)
GOPlot (1)
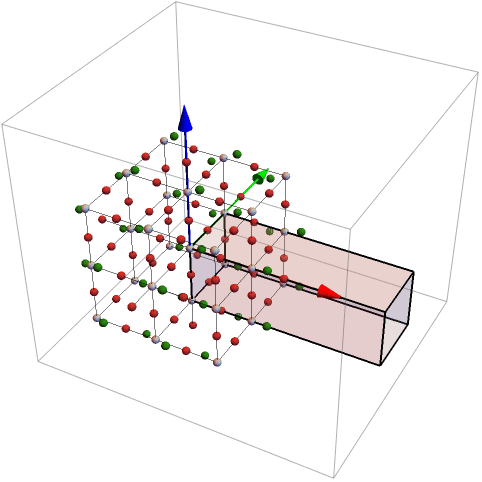
If it is set to True, the result is plotted and the also the sub-cluster is given.
Now the atoms found in the parallelepiped are plotted by means of GTPlotCluster.